Extracting raw waveforms¶
Extracting datasnippets from ImecDataset¶
It is often useful to extract small windows of time from the raw data on each channel. Given a list of sample indices times you want to extract a window of time around, and a window = [offsetPre, offsetPost] of relative samples to collect around those times, you can do the following:
>> times = 1e5:1e5:1e6;
>> window = [-1000, 999]; % 1000 samples before plus 999 samples after
>> snippetSet = imec.readAPSnippetSet(times, window)
snippetSet =
SnippetSet with properties:
data: [384×2000×10 int16] % nChannels x nTimepoints x nSnippets snippet data
cluster_ids: [10×1 uint32]
channel_ids_by_cluster: [384×1 uint32] % nChannels x nClusters channel ids, which channel ids were extracted for each cluster
unique_cluster_idx: 1
sample_idx: [10×1 uint64]
trial_idx: [0×1 uint32]
window: [-1000 999]
valid: [10×1 logical]
channelMap: [1×1 Neuropixel.ChannelMap]
scaleToUv: 2.3438
fs: 30000
nChannels: 384
nTimepoints: 2000
nSnippets: 10
nClusters: 1
data_valid: [384×2000×10 int16]
Extracting waveforms via KilosortDataset¶
With this functionality in place, it is very easy to extract raw waveforms from the original ImecDataset guided by a KilosortDataset to provide the spike times. From a KilosortDataset, you can request waveforms based on one of:
spike_idx: index directly intospike_timesspike_times: find spikes that match times inspike_timescluster_ids: find all spikes from a given cluster. Often, you would also specify'num_waveforms'to cap the total number of waveforms per cluster via random sample, unless you want every spike for those clusters
By default, waveforms will be extracted for every channel on the probe, unless you request specific channel_ids_by_cluster as a nClusters x nChannels matrix, or specify best_n_channels which will pick the best channels separately for each cluster.
snippetSet = ks.getWaveformsFromRawData('cluster_ids', cluster_id, ...
'num_waveforms', 50, 'best_n_channels', 20, 'car', true);
snippetSet.plotAtProbeLocations('alpha', 0.8);
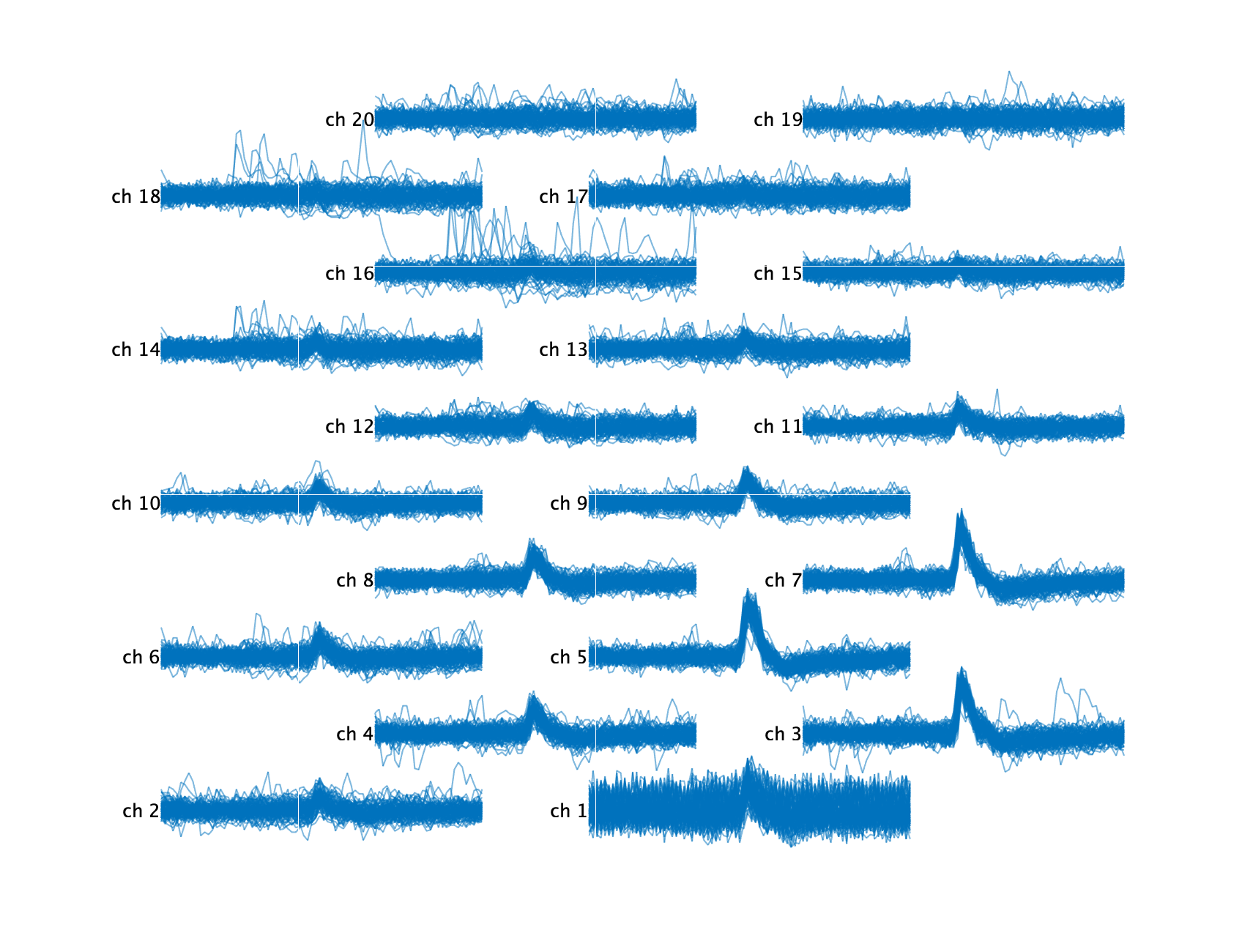
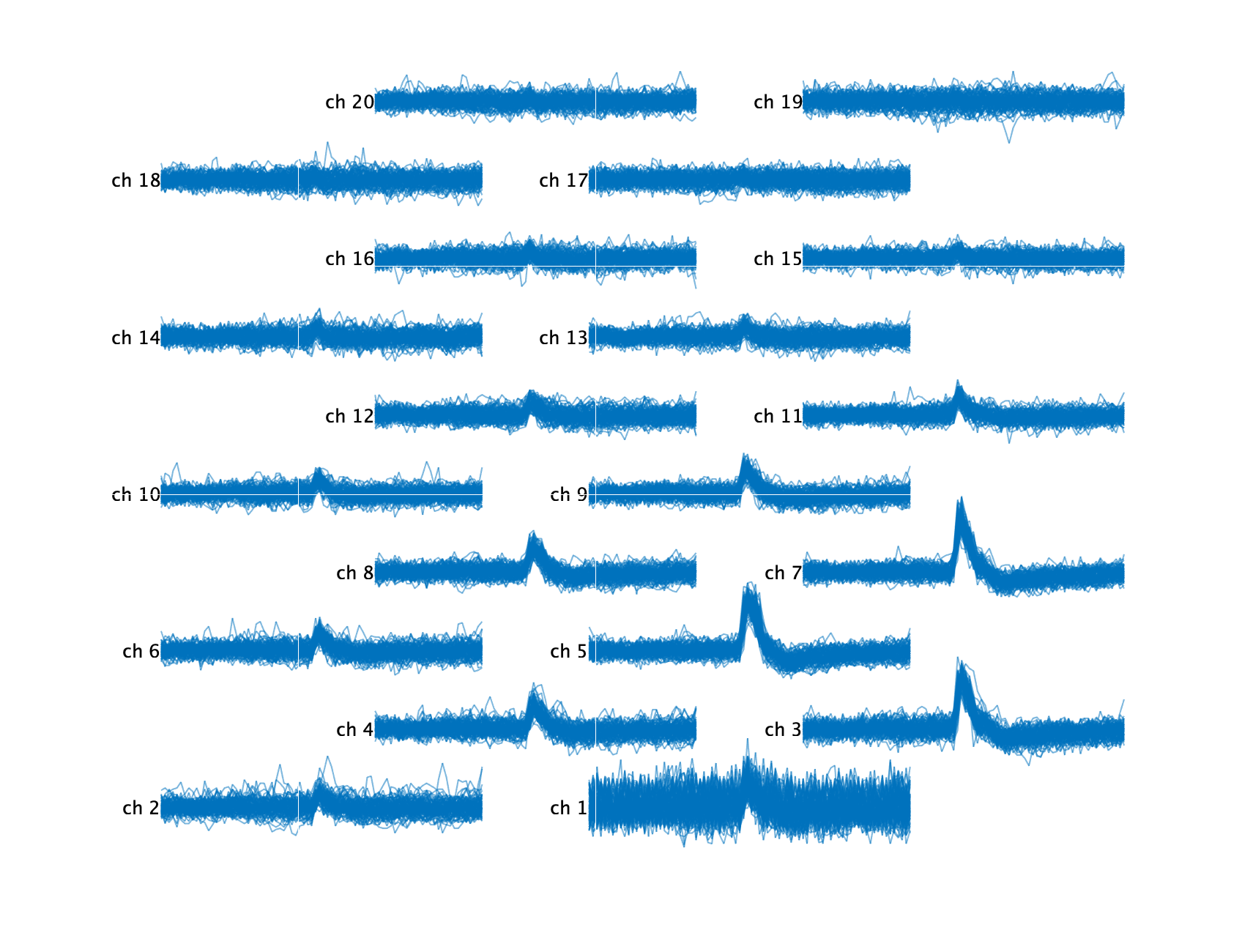
As you can see, there are other waveforms in the background corrupting these extracted waveforms. We can use the other Kilosort detected spike times and their respective templates to automatically clean these waveforms by subtracting the appropriate template at the appropriate times.
snippetSet = ks.getWaveformsFromRawData('cluster_ids', cluster_id, ...
'num_waveforms', 50, 'best_n_channels', 20, 'car', true, ...
'subtractOtherClusters', true);
snippetSet.plotAtProbeLocations('alpha', 0.8);
Extracting waveforms via KilosortTrialSegmentedDataset¶
If you have already segmented the data into trials, you might wish to extract these waveforms for specific trials or even specific time windows within a trial, e.g. to check for behaviorally-triggered probe movement.
For example, this will extract all spikes that occurred in the first 50 ms of each trial:
cluster_ind = seg.lookup_clusterIds(cluster_id);
mask_by_trial = cell(seg.nTrials, 1);
for iTrial = 1:seg.nTrials
mask_by_trial{iTrial} = seg.spike_times_ms_rel_start{iTrial, cluster_ind} < 50;
end
snippetSet = seg.getWaveformsFromRawData('mask_cell', mask_by_trial, 'cluster_id', cluster_id, ...
'best_n_channels', 20, 'subtractOtherClusters', true);